Since their discovery in sinoatrial node cells and neurons in the late 1970s and early 1980s, Hyperpolarization-Activated Cyclic Nucleotide-Gated (HCN) channels have sparked continuing interests. The use of Alomone Labs antibodies directed towards HCN channels provides an extensive research tool for studying the importance of these channels regarding structure-function correlations, elucidating their location in various tissues, identifying interaction sites with various proteins and elucidating mechanisms underlying pathology and diseases caused by specific defects in HCN channels.
Introduction
HCN channels comprise a small subfamily of proteins within the superfamily of pore-loop cation channels. In mammals, the HCN channel family includes four members (HCN1-4) that are expressed in the heart and in the nervous system. Hyperpolarization-activated cation currents produced by HCN channels (Ih, If or Iq), contribute to a wide range of physiological functions including cardiac and neuronal pacemaker activity, the setting of resting potentials, dendritic integration of synaptic transmission and learning8,42. Throughout this article If refers to cardiac currents and Ih refers to non-cardiac currents.
HCN channels are unique in three ways: i) channel gating by membrane hyperpolarization. In general, Ih currents activate with hyperpolarizing steps to potentials negative to -50 to -60 mV. Unlike most voltage-gated currents, Ih does not display voltage-dependent inactivation. ii) Ih is regulated by cyclic nucleotides (cAMP). iii) Ih is a mixed cation current that is carried by both K+ and Na+ under normal physiological conditions8.
In the heart, HCN1 and HCN2 are somewhat expressed, while HCN4 is the most abundant subunit. In the mammalian brain, all four HCN isoforms are expressed and are particularly wellcharacterized in the hippocampus. The four HCN subunits can form either homo or hetero tetrameric structures that are arranged around the centrally located pore. HCN channels can also coassemble with auxiliary subunits to form channels with modified properties8.
Developmental Regulation
Although If is an important component of spontaneous activity in the embryonic heart, the contribution of this current to the differentiation of embryonic stem (ES) cells into cardiac cells remains unknown.
Changes in the expression and function of HCN channels during the differentiation of cardiac precursor cells derived from mouse ES cells were examined. HCN1 and HCN4 were predominantly expressed using Anti-HCN1 (#APC-056) and Anti-HCN4 (#APC-052) antibodies along with If currents which were predominantly observed in cardiac precursor cells on day 1552 (Figure 1).
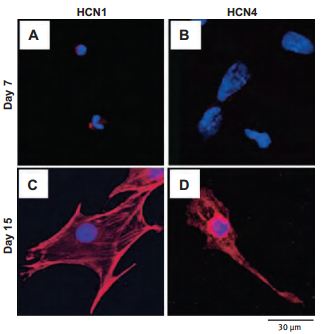
Cells from day 7 (A and B) and day 15 (C and D) of differentiation were sorted by FACS, then fixed and stained using Anti-HCN1 (A and C) and Anti-HCN4 (B and D) antibodies (#APC-056 and #APC-052, respectively) as indicated (red). Nuclei were stained using DAPI (blue).
Adapted from reference 52 with permission of Biomedical Research.
Another work used spontaneously beating cells derived from mouse embryonic stem cells (mESCs) in culture. Using Anti-HCN2 (#APC-030) and Anti-HCN3 (#APC-057) antibodies it was demonstrated that the proportion and density of If increased during development. Results from western blot analysis are consistent with the presence of HCN2 and HCN3 suggesting that If confers regular and faster rhythmicity in mESCs40.
While all four HCN channels are expressed in the cerebellum, the contribution of the individual subunits to the proposed cerebellar functions of Ih is currently unknown. The presented study investigated the developmental profile, the selective distribution pattern and the cellular and subcellular localization of HCN1 in the cerebellum22 (Figure 2). Using Anti-HCN1, the authors found that HCN1 is indeed expressed in the rat cerebellum at P60 (postnatal day 60) (Figure 2A). In an attempt to study the expression of HCN1 during development, rat brain membranes were extracted (P0-P60) and subjected to western blot analysis. HCN1 expression became evident at P5 and peaked at P15 (Figure 2B). Confocal microscopy of immunohistochemical studies showed similar results and suggested that HCN1 is localized in axon terminals of cerebellar basket cells (Figure 2C). HCN1 expression is also developmentally regulated in the integrated compartments of the layer 5 pyramidal neurons as determined in immunohistochemical analysis using Anti-HCN15. HCN2 expression was also found to be developmentally regulated in the hippocampus using its respective Alomone Labs antibody48.
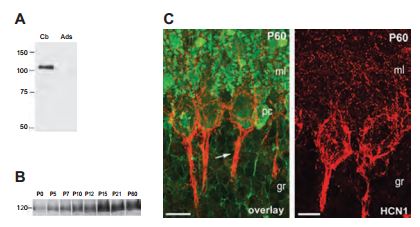
Immunoblot analysis of crude membrane preparations from adult (A) and developing cerebellum (B) with Anti-HCN1 antibody (#APC-056). A) No immunoreactivity was detected after adsorption of the Anti-HCN1 with the immunizing negative control antigen. B) During the postnatal development, HCN1 was barely detected at birth and increased steadily through to P15 and reached a plateau at P21. C) Immunohistochemistry of cerebellar cortex sections with Anti-HCN1. Most of the labeling of HCN1 at P60 is confined to basket and pinceau-like structures (arrow) around Purkinje cells. Scale bars, 20 μm.
Adapted from reference 22 with permission of Blackwell Publishing Ltd.
Distribution and Function Studies
The distribution of HCN channels in the SAN was investigated using histology and immunolabeling applications. Anti-HCN1, Anti-HCN2 and Anti-HCN4 antibodies revealed that HCN4 is the only HCN isoform detectable in SAN cells. Strands of HCN4-positive nodal cells also protruded into the atrial muscle (Figure 3). The data suggest that this specialized interface between the SAN and the surrounding atrium may be necessary for the SAN to drive the more hyperpolarized atrial muscle21. In a different study, immunolabeling using Anti-HCN4 antibody demonstrated that the cells in the central SAN area are predominantly distributed in islets interconnected by cell prolongations (Figures 4A and 4B) and single-cell HCN4 labeling suggested sites of channel clustering (Figure 4C). It was therefore proposed that expression of HCN4 is an appropriate tool to map and identify the cardiac SAN pacemaker region9.
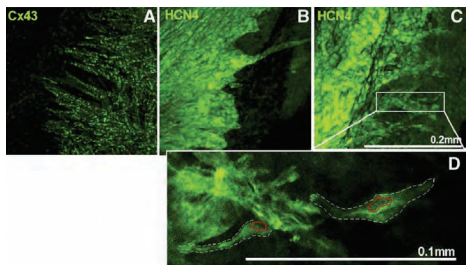
Cx43 (A) and HCN4 (B, C) labeling using Anti-HCN4 antibody (#APC-052) at the border of the SAN (Cx43-negative/HCN4-positive) and the atrial muscle (Cx43-positive/HCN4-negative) towards the atrial septum. Projection images from different preparations are shown (A-C). D) High magnification image of HCN4 labeling from the boxed region in (C). Two SAN cells are highlighted by the white dashed lines and their nuclei are highlighted by the red dotted lines. Scale bars, 200 μm (A-C) and 100 μm (D).
Adapted from reference 21 with permission of Oxford University Press.
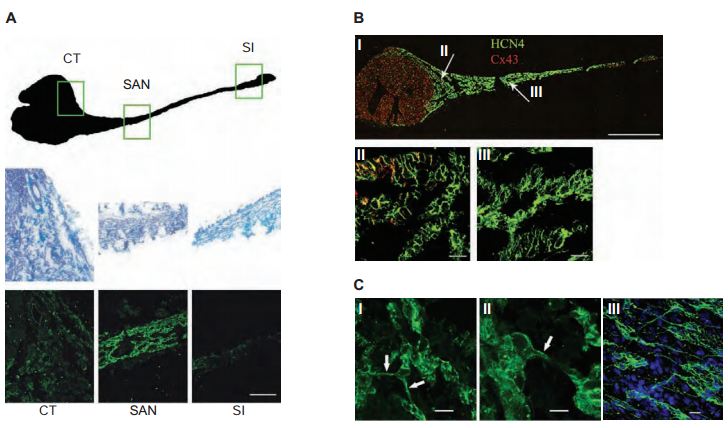
A) Regions of nodal tissue investigated (top). The CT, SAN and SI regions delimited by the boxes are enlarged using masson-trichrome staining of the selected regions of the intercaval area (middle panels). Immunofluorescence staining using Anti-HCN4 antibody (#APC-052) confirms that the HCN4 signal arising from primary pacemaker cells (SAN) is higher than that from other more peripheral regions (bottom panels). Scale bars, 20 μm. B) HCN4 (green) and Cx43 (red) immunolabeling of the rabbit intercaval area (I); enlargements corresponding to the regions identified by arrows in panel (I) are shown in panels (II) and (III). HCN4 signal, detected with Anti-HCN4 clearly labels the whole SAN region, while Cx43 is only detectable outside the node or in its peripheral regions (II, III). For both proteins the signal distribution shows a selective labeling of the peripheral and central SAN areas. Scale bars, (I) 1 mm; (II, III) 20 μm. C) Cellular organization of the central region of the SAN. High resolution images of sections obtained in the central area of the node (I, II). Cells are organized in groups (islets) interconnected by thin cytoplasmic bridges (arrows). Whole-tissue labeling with Anti-HCN4 confirms the cellular organization in islets and bridges (III). Scale bars, 10 μm.
Adapted from reference 9 with permission of Elsevier.
The distribution of HCN4 using Anti-HCN4 was studied in the center, periphery and atrial muscles of the mouse SAN pacemaker cell area. In addition to HCN4, two Na+ channel isoforms were found important for proper SAN function: neuronal voltage-gated Na+ channel (putatively Nav1.1) and cardiac Nav1.5 isoforms18.
The possibility that both HCN4 and HCN1 isoforms contribute to the native If in SAN cells by forming heteromeric channels was explored in HEK293 cells, using Anti-HCN4 antibody. Co-transfected (HCN4 + HCN1) and various chimeric constructs with N- and C- termini of either channel were compared with those of the native If current. Results indicate that HCN1 and HCN4 may contribute to native If channels, but a ‘context’-dependent mechanism is also likely to modulate the channel properties in native tissues3. MiRP1 was also found to contribute to modulate these currents (determined using Anti-KCNE2 (MiRP1) antibody (#APC-054)).
The co-assembly of HCN2 and HCN4 in live CHO cells was studied using bioluminescence resonance energy transfer (BRET2), a novel approach for studying tetramerization of ion channel subunits. Together with results from electrophysiological and imaging approaches, BRET2 data showed that HCN2 and HCN4 subunits self-assemble and co-assemble with equal preference. The colocalization of HCN2 and HCN4 in the embryonic mouse heart in immunohistochemistry studies was also demonstrated using Anti-HCN2 and Anti-HCN4 antibodies. Together, these data support the formation of HCN2-HCN4 heteromeric channels in native tissues51.
It was recently discovered that constitutively active Src tyrosine kinase can enhance HCN4 channel activity by binding to the channel. In order to investigate the mechanism of HCN4 modulation by Src, the effects of PP2, a selective inhibitor of Src, on HCN4 was studied by using Anti-HCN4 antibody in immunoprecipitation and western blot analyses in HEK 293 cells. Results suggest that Src enhances HCN4 currents via Tyr53119.
Cysteine scanning substitutions were introduced into the descending portion of the HCN1 P-loop. Although expressed (as demonstrated by western blot analysis using Anti-HCN1), all mutated channels were devoid of functional currents. It was concluded that specific pore residues are important determinants of the structure-function properties of HCN channels in general6. Cysteine scanning substitutions were also introduced in the P-S6 linker of HCN1. The novel residues identified display extracellular accessibility which is steric and state dependent. These data provide the first functional evidence consistent with a pore-to-gate model for HCN channels45.
Alomone Labs Anti-HCN2 and Anti-HCN4 antibodies were both used in western blot and immunoprecipitation experiments and revealed that in the myocardium, the molecular mass of HCN2 is significantly lower, lacking the C-terminus cAMP binding domain. In heterologous cells, the C-terminal-truncated HCN2 protein co-assembles with HCN4 to form functional heteromeric HCN channels, which activate faster than homomeric HCN2 or HCN4 channels, and display properties similar to endogenous myocardial If channels. Taken together, the results suggest that functional myocardial If channels reflect the heteromeric assembly of HCN2 and HCN4 and furthermore, that HCN4 underlies the cAMP-mediated regulation of cardiac If channels53. Heteromeric association between HCN2 and HCN4 was also demonstrated in Xenopus oocytes with Anti-HCN2 and Anti-HCN4. These structures also displayed faster activation kinetics than the homomeric channels55.
Another work directly investigated the role of cAMP-dependent regulation of HCN channels in SAN activity. Mice with heart-specific and inducible expression of a human HCN4 mutation (abolishing the cAMP-dependent regulation) were generated. The expression of HCN4 was assessed by western blot analysis, using Anti-HCN4 antibody. This mutant mouse model suggests that cAMP-mediated regulation of HCN determines basal and maximal heart rates but does not play an indispensable role in heart rate adaptation during physical activity2.
Another work investigated biological pacemakers (BPM) implanted in canine, which function competitively with electronic pacemakers (EPM). The engineered BPM (BPM/EPM tandem) was implanted in rats mutated in HCN2 (thereby mimicking arrhythmia) and found that the BPM/EPM tandems confer advantage over either approach used on its own. In this study, Anti-HCN2 antibody was used to show that mutated rat cardiomyocytes express much lower levels of HCN2 than wild-type10.
In the ear, the axis of the cochlea encloses the cell bodies of the spiral ganglion cells (SGCs). Type I and type II SGCs are responsible for afferent innervation of the inner and outer hair cells of the cochlea respectively. HCN channels and Ih currents have been identified in type I SGCs and Ih currents have been observed in type II SGCs11,14,29. Recently, all four HCN4 subunits were identified in both types of SGCs7 . Western blot analysis, using whole spiral ganglion lysates was done in order to verify the specificity of the antibodies. As shown in Figure 5, Anti-HCN2 and Anti-HCN3 detected specific bands (Figure 5A). Fluorescent immunohistochemistry applied on cochlear free-floating preparations as well as wax-embedded tissues showed that HCN2 and HCN3 immunoreactivities are present on the cell bodies as well as the processes (Figures 5B and 5C).
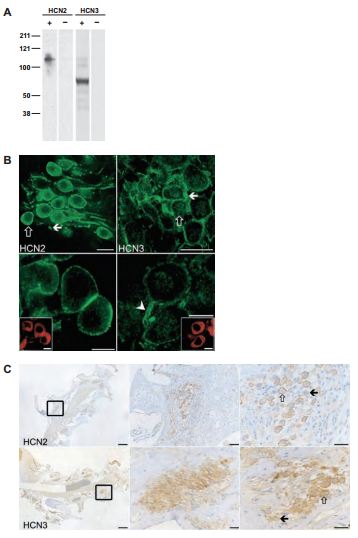
A) Western blot analysis of whole spiral ganglion tissue preparation isolated from guinea pigs. Lanes with + signs demonstrate lysates from tissue preparations reacted with Anti-HCN2 (#APC-030) and Anti-HCN3 (#APC-057) antibodies. Lanes with – signs indicate results of preadsorbed control experiments. B) The results of HCN2 and HCN3 specific immunoreactivities from cochlear free-floating tissues (upper left and right panels respectively) and their respective high magnification images (lower panels). Scale bars, 20 μm (upper panels); 10 μm (lower panels). C) Wax-embedded cochlear tissues were subjected to Anti-HCN2 and Anti-HCN3 treatment (upper and lower panels respectively). The rectangle in the left panels indicates an example of a cellular nest accommodating the cell bodies of the SGCs. Middle and right panels represent higher magnification. Open and filled arrows represent examples of strongly and weakly labeled cells respectively. Scale bars, 250 μm (upper and lower left panels); 50 μm (upper middle panel); 25 μm (upper right panel and lower middle and right panels).
Adapted from reference 7 with permission of Elsevier.
Pyramidal cells are one of the most prominent cell types in the dorsal cochlear nucleus. Immunocytochemical labeling identified HCN1, HCN2 and HCN4 from acutely isolated dorsal cochlear nuclear neurons using Alomone Labs antibodies and all three channels were found expressed by pyramidal cells35. In addition, HCN1 distribution was found to overlap with that of Kv1.1 in cellular regions of the cochlear nucleus34.
The medial septum/diagonal band (MS/DB) iis a region in the brain which initiates and/or modulates the hippocampal theta rhythm, important for memory and learning46,49. Although in situ hybridization studies did show expression of HCN1, 2, and 4 in MS/DB30,41, the extent of the overlapping expression of these channels is not clear. In accordance with the mRNA detection of all three channels in MS/DB30,41, immunohistochemical analysis using Anti-HCN1, 2, and 4 antibodies, showed that all three channels are expressed in MS/DB nuclei31. HCN1 detection was strong in somatic and proximal dendritic surface membrane of neurons. Expression of HCN2 was similar to that of HCN1 but mostly cytoplasmic. HCN4 on the other hand had a more widespread staining.
Ih currents are also present in the soma of sensory neurons that do not exhibit any of the pacemaker or oscillatory potentials. The aforementioned neurons were used to study the expression of HCN1, 2, and 4, using their respective Alomone Labs antibodies in immunohistochemical studies12. HCN1 immunoreactivity was confined to only a subpopulation of nodose neurons, whereas HCN2 and HCN4 were expressed in all neurons of the nodose ganglion. Inhibiting Ih in the sensory neurons rendered the neurons more excitable and no role for oscillatory behavior was found.
While HCN1-4 expression has been described in the thalamus, HCN2 and HCN4 are the major isorforms in the glutamatergic ventrobasal complex (VB) and the GABAergic reticular thalamic nucleus (RTN)33. However, their exact compartmental location is still unclear. Immunohistochemical studies with AntiHCN2 and Anti-HCN4 showed that they are individually expressed in both RTN and VB neurons, and mostly overlap in the VB1. HCN2 but not HCN4 was distributed in dendritic spines in both RTN and VB neurons, important in the regulation of neuronal excitability and synaptic plasticity1. The role of HCN2 in RTN neurons was therefore investigated54. Ih was abolished in Hcn2-/ – cells strongly suggesting that HCN2 is the major contributor to Ih currents in RTN. Confocal microscopy images demonstrated punctate and sparse somatic labeling of HCN2 immunoreactivity in wild-type cells and was absent in all cellular compartments in Hcn2-/- cells. On the other hand, Hcn2 deletion did not affect HCN4 somatic immunoreactivity (Figure 6). The electrophysiological data in the study strongly suggested that Ih mediated by HCN2 acts as a leak current path in order to “turn off” excitatory inputs. This implies that HCN2 and glutamate receptor subunit GluR4 may be in close proximity, as they are both expressed in RTN neurons54.

A) High magnification images showing triple immunofluorescent labeling for parvalbumin (blue), MAP2 (green), and HCN2 (red) in Hcn2+/+ (left panel) and Hcn2-/- (right panel) in RTN neurons using Anti-HCN2 antibody (#APC-030). B) Immunostaining with Anti-HCN4 antibody (#APC-052) shows prominent labeling in both Hcn2+/+ and Hcn2-/- RTN neurons. Neither HCN2 nor HCN4 overlap with MAP2. Scale bars, 5 μm.
Adapted from reference 54 with permission of The Society for Neuroscience.
All four HCN channels have unique but overlapping expression patterns in the CNS. The distribution of HCN1 in the brainstem and spinal cord was examined in immunohistochemical studies28. At all levels of the spinal dorsal horn, HCN1 immunoreactivity was quite dense in laminae III. In the brainstem, HCN1 expression was included in neurons in sensory pathways as well as motoneurons.
HCN channels are abunddantly expressed in DRGs. A study was initiated to see whether HCN2 is localized in the DRG soma or whether it is transported from the soma to the central axons of the DRG neurons that terminate in the spinal dorsal horn4 (Figure 7). HCN2 was located in primary sensory neurons in the superficial spinal dorsal horn of rats, more specifically in laminae I and outer laminae II (Figure 7A). Furthermore, dorsal rhyzotomy completely abolished the immunoreactivity of HCN2 (Figure 7B), strongly suggesting that HCN2 is confined to axon terminals of nociceptive primary afferents. To further expand this notion, HCN2 was found to colocalize with CGRP, a neuropeptide expressed by peptidergic nociceptive primary afferents (Figure 7C). Having gained insight on the localization of HCN2 in nociceptive primary afferents, a subsequent study was done to investigate the functional significance of HCN2 in nociceptive primary afferents36. HCN2 colocalized with Substance P (SP), suggesting that HCN2 may be involved in pain transmission from SPcontaining nociceptive primary afferents to spinal neurons (Figure 7D).
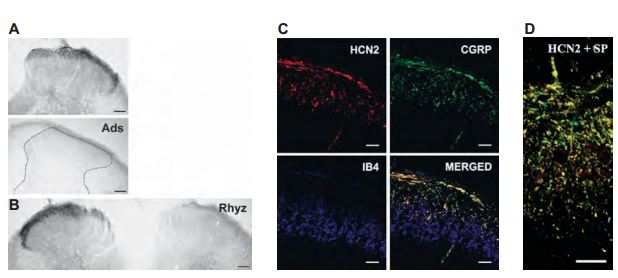
A) Micrographs showing HCN2 immunoreactivity using Anti-HCN2 antibody (#APC-030) in laminae I and outer II in spinal dorsal horn (upper panel). Adsorption of AntiHCN2 to the negative control antigen completely abolished immunostaining. B) Dorsal rhyzotomy abolished the immunostaining from the dorsal horn. C) Three images from a single superficial spinal dorsal horn section was labeled for HCN2 (red, upper left panel), CGRP (green, upper right panel), and IB4-binding (blue, lower left panel). The superimposed image (lower right panel) indicates a strong colocalization between HCN2 and CGRP in laminae I and outer II (yellow color). IB4-binding is completely segregated from both HCN2 and CGRP immunoreactivity. D) Micrographs showing colocolization between HCN2 (red) and substance P (SP, green). Scale bars, 100 μm (A and B); 20 μm (C); 10 μm (D).
Adapted from references 4 and 36 with permission of Blackwell Publishing Ltd.
Afferent pathways innervating the urinary bladder consist of dorsal root ganglion (DRG) neurons (medium- and small-sized cell populations). Since Ih currents have been identified in various peripheral sensory neurons, the expression of HCN channels in bladder afferent neurons from L6-S1 spinal cord DRGs was examined25. Using Alomone Labs Anti-HCN1, 2 and 4 antibodies, HCN2 showed the most intense immunoreactivity in DRGs. In contrast to HCN1, HCN2 is found in laminae I and II of the dorsal horn, whereas HCN1 is found in laminae III28 as mentioned above.
Modulating Ih and Ik-K-leak (mediated by TASK K+ channels) plays an important role in determining the resting potential. Thalamocortical relay (TC) neurons are an ideal model for understanding the interaction between Ih and Ik-K-leak, and resting membrane potential. Immunohistochemical studies showed that 100% of TC neurons are positive for both HCN2 and TASK3 and that both currents contribute to the resting membrane potential26.
HCN and Protein-Protein Interactions
Channel activity depends in many cases on the interaction with auxiliary subunits and scaffolding proteins. HCN channel subunits are believed to be modulated by additional regulatory proteins, which only recently have been identified.
MinK-related protein (MiRP1 or KCNE2) was previously found to interact with the HCN family of pacemaker channels, and to alter channel gating in heterologous expression systems. In order to evaluate the effect of MiRP1 on HCN expression and function in a physiological context, overexpressed HA-tagged MiRP1 (HA-MiRP1) and HCN2 expressed in neonatal rat ventricular myocytes was investigated. Coimmunoprecipitation experiments using AntiHCN2 antibody demonstrated that HA-MiRP1 and HCN2, as well as endogenous MiRP1 and HCN2, co-assemble in ventricular myocytes. The results indicate that MiRP1 acts as a β subunit for HCN2 and alters channel gating in cardiac cells39.
A protein-protein interaction between HCN2 and the K+ channel regulator protein 1 (KCR1) was revealed. Coimmunoprecipitation experiments, using Anti-HCN2 antibody, showed that KCR1 and HCN2 associate. The results further showed that KCR1 modulates IHCN2/If channel gating in cardiac cells27. In addition, usage of Anti-HCN2 and Anti-HCN4 antibodies showed that both HCN2 and HCN4 isoforms co-distribute with the adapter protein SAP97, an important component in the SAN. HCN4, but not HCN2, also co-distributes with the post-synaptic marker β-catenin, thus identifying diverse organized domains within this tissue. Together, their data suggest that SAP97 contributes to isoform specific organization of HCN channels within specific domains in the SAN37.
In the brain, a yeast two hybrid screen showed that HCN2 interacts with Tamalin, a scaffold protein, through the PDZ-binding motif15. These two proteins indeed interact as they could both be immunoprecipitated using Anti-HCN2 antibody. In addition, immunoprecipitating rat brain membrane showed that along with HCN2, S-SCAM and Mint2, two neuronal scaffold proteins, are both pulled down with the channel. These scaffold proteins may unequivocally direct proper localization, influence trafficking and signal transduction of HCN channels in neuronal cells. In a different yeast two hybrid screen, Filamin A, also a scaffold protein was found to interact with the C-terminal of HCN113. in vivo interaction was also demonstrated using Anti-HCN1 antibody. Filamin A may be involved in clustering HCN113. Pex5R was first identified via a yeast two hybrid screen as an HCN interacting protein43. In a recent study, Pex5R was also identified as an HCN interacting protein when Anti-HCN2 antibody was used to immunoprecipitate the channel from membrane fractions that were prepared from rat brain57. The role of Pex5R is actually to slow the time course of HCN channel activation by allosterically hindering cAMP binding in the brain.
HCN, Disease and Channelopathies
The overexpression of If has been functionally demonstrated in ventricular myocytes in failing human hearts. The molecular basis however of If overexpression in human cardiac disease is still unknown.
Little is known about the expression of HCN channels in HCN remodeling by diseases, such as congestive heart failure (CHF), associated with disturbances of cardiac rhythm. The expression of HCN1, HCN2 and HCN4 in normal dogs was assessed compared to dogs subjected to 2-week ventricular tachypacing-induced CHF. Western blot and immunohistochemistry analyses, using Anti-HCN2 and Anti-HCN4 antibodies showed that HCN2 and HCN4 expression are greater at both protein and mRNA levels in the SAN than in the right atrium (RA). In the RA, HCN4 expression but not that of HCN2 is significantly upregulated by CHF. The overall findings provide new information about the molecular basis of normal and disease-related impairments of cardiac impulse formation56.
A missense mutation in the ion channel pore domain of HCN4 is associated with inherited sinus bradycardia. Expression of the wild-type and mutant HCN4 channel was assessed in HEK293 cells using Anti-HCN4. Results show that cells transfected with mutated HCN4 express much lower levels than those transfected with wildtype. Despite its critical location, this mutation carries a favorable prognosis without the need for pacemaker implantation during long-term follow-up32.
The mechanisms involved in the impairment of arterial baroreflex in type-1 diabetes (T1D), have yet to be deciphered. The nodose ganglion (NG) contains the cell bodies of the aortic baroreceptor (AB) neurons which express HCN channels important for regulating the cell excitability. Immunofluorescence and western blot analyses using Anti-HCN1 and Anti-HCN2 antibodies showed that the expression of HCN1 and HCN2 is higher in T1D rats compared to wild-type. The results indicate that HCN channels contribute to the decreased excitability of AB neurons in T1D rats20.
The genetic and molecular mechanisms underlying human sinus node dysfunction (SND) are limited. Penetrant and severe SND was mapped to the human ANK2 (ankyrin-B/AnkB) locus. AnkB is essential for normal membrane organization of SAN cell channels and transporters, and is also required for physiological cardiac pacing. Anti-HCN4 antibody was used as a SAN marker in immunohistochemistry in AnkB+/- and AnkB+/+ SAN cells from mouse sections (Figure 8). Their findings reveal abnormal channel targeting (but not HCN channels) with SND and highlight the critical role of local membrane organization for SAN excitability17.
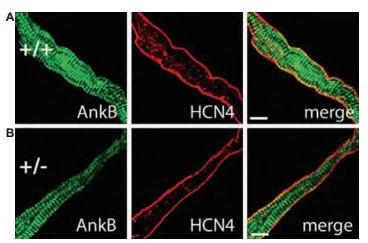
A) and B) Confocal imaging of SAN cells from AnkB+/+ (wild type) and AnkB+/- mice. SAN cells were immunolabeled, using Anti-HCN4 antibody (#APC-052). Wild type and AnkB+/- SAN cells displayed no difference in the expression or localization of HCN4. Scale bars, 10 μm.
Adapted from reference 17 with permission of The National Academy of Sciences of the USA (copyright 2008).
Enhanced abnormal automaticity of ventricular cells contributes to hypertrophic arrhythmias. The expression of HCN2 and HCN4 is reportedly increased in hypertrophic and failing hearts, contributing to arrhythmogenesis. A study on post-transcriptional regulation of HCN2 and HCN4 by microRNAs was initiated in the rat model of left ventricular hypertrophy. The use of Anti-HCN2 and Anti-HCN4 antibodies in western blot analysis revealed the robust increase in HCN2 and HCN4 levels in a rat model of ventricular hypertrophy, accompanied by pronounced reduction of microRNA levels. Downregulation of HCN2 and HCN4 targeted microRNAs contributes to the re-expression of HCN2/HCN4 and thereby the electrical remodeling process in hypertrophic hearts24.
The expression of HCN was assessed in ventricular and atrial samples from normal or failing hearts explanted from patients with endstage ischemic cardiomyopathy. HCN2 and HCN4 were detected in human myocardium, using Anti-HCN2 and Anti-HCN4 antibodies in western blot studies and their levels were significantly higher in failing ventricles and atrial samples. HCN upregulation likely contributes to increased If and may play a role in ventricular and atrial arrhythmogenesis in heart failure47.
Although absence epilepsy has a genetic origin, evidence from an animal model suggests that seizures are sensitive to environmental manipulations. Manipulation of the early rearing environment of epileptic rats leads to a pronounced decrease in seizure activity later in life. The alterations in seizure activity between rats reared differently might be correlated with changes in Ih and its channel subunits. Western blot analysis of the somatosensory cortex revealed a long term increase in Ih and HCN1 expression specifically (using Anti-HCN1, 2, and 4 antibodies), in neonatal handled and maternal deprived44. An extended work showed, that HCN1 expression undergoes a rapid decline (along with a decline in Ih), preceding the onset of seizures16. Epileptic rat cortex demonstrated weak immunolabeling of HCN1 and Ih currents as opposed to control rat cortex. Abnormal activity in DRG function may be involved in higher pain sensitivity or spontaneous pain sensation among some patients. As HCN channels have been identified in DRGs, their involvement was investigated regarding pain sensation23. Immunohistochemistry sections demonstrated that all four HCN channels are expressed in peripheral sensory terminals within plantar skin sections (Anti-HCN2 and Anti-HCN4 were both purchased from Alomone Labs). The data strongly suggested that expression and modulation of Ih in sensory organs and nerve fibers in the periphery play a role in somatosensory processing and contribute to both spontaneous pain and tactile allodynia.
The preoptic area (PO) and the anterior hypothalamus (AH) of the rostral hypothalamus are important integrative centers in the regulation of body temperature50. Neurons are classified as temperature sensitive (warm or cold), temperature insensitive and silent. Immnunohistochemical studies showed that HCN2 and HCN4 labeling are found (using Anti-HCN2 and Anti-HCN4) in the PO/AH at all levels and are important for their activity50.
References
- Abbas, S.Y. et al. (2006) Neuroscience 141, 1811.
- Alig, J. et al. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12189.
- Altomare, C. et al. (2003) J. Physiol. 549, 347.
- Antal, M. et al. (2004) Eur. J. Neurosci. 19, 1336.
- Atkinson, S.E. and Williams, S.R. (2009) J. Neurophysiol. 102, 735.
- Au, K.W. et al. (2008) Am. J. Physiol. 294, C136.
- Bakondi, G. et al. (2009) Neuroscience 158, 1469.
- Biel, M. et al. (2009) Physiol. Rev. 89, 847.
- Brioschi, C. et al. (2009) J. Mol. Cell. Cardiol. 47, 221.
- Bucchi, A. et al. (2006) Circulation 114, 992.
- Chen, C. (1997) Hear. Res. 110, 179.
- Doan, T.N. et al. (2004) J. Neurosci. 24, 3335.
- Gravante, B. et al. (2004) J. Biol. Chem. 279, 43847.
- Jagger, D.J. and Housley, G.D. (2003) J. Physiol. 552, 525.
- Kimura, K. et al. (2004) Genes Cells. 9, 631.
- Kole, M.H.P. et al. (2007) J. Physiol. (London) 578, 507.
- Le Scouarnec, S. et al. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 15617.
- Lei, M. et al. (2004) J. Physiol. 559.3, 835.
- Li, C.H. et al. (2008) Am. J. Physiol. 294, C355.
- Li, Y.L. et al. (2008) Cardiovasc. Res. 79, 715.
- Liu, J. et al. (2007) Cardiovasc. Res. 73, 729.
- Lujan, R. et al. (2005) Eur. J. Neurosc. 21, 2073.
- Luo, L. et al. (2007) Neuroscience 144, 1477.
- Luo, X. et al. (2008) J. Biol. Chem. 283, 20045.
- Matsuyoshi, H. et al. (2006) Brain Res. 1119, 115.
- Meuth, S.G. et al. (2006) J. Neurophysiol. 96, 1517.
- Michels, G. et al. (2008) PLoS One 3, e1511.
- Milligan, C.J. et al. (2006) Brain Res. 1081, 79.
- Mo, Z.L. and Davis, R.L. (1997) J. Neurophysiol. 78, 3019.
- Monteggia, L.M. et al. (2000) Brain Res. Mol. Brain. Res. 81, 129.
- Morris, N.P. et al. (2004) Brain. Res. 1006, 74.
- Nof, E. et al. (2007) Circulation 116, 463.
- Notomi, T. and Shigemoto, R. (2004) J. Comp. Neurol. 471, 241.
- Oertel, D. et al. (2008) Neuroscience 154, 77.
- Pal, B. et al. (2003) Cell. Mol. Life Sci. 60, 2189.
- Papp, I. et al. (2006) Eur. J. Neurosci. 24, 1341.
- Peters, C.J. et al. (2009) J. Mol. Cell. Cardiol. 46, 636.
- Proenza, C. et al. (2002) J. Biol. Chem. 277, 5101.
- Qu, J. et al. (2004) J. Biol. Chem. 279, 43497.
- Qu, Y. et al. (2008) J. Physiol. 586, 701.
- Santoro, B. et al. (2000) J. Neurosci. 20, 5264.
- Santoro, B. and Tibbs, G.R. (1999) Ann. N. Y. Acad. Sci. 868, 741.
- Santoro, B. et al. (2004) J. Neurosci. 24, 10750.
- Schridde, U. et al. (2006) Eur. J. Neurosci. 23, 3346.
- Siu, C.W. et al. (2009) J. Membr. Biol. 230, 35.
- Stewart, M. and Fox, S.E. (1990) Trends Neurosci. 13, 163.
- Stillitano, F. et al. (2008) J. Mol. Cell. Cardiol. 45, 289.
- Surges, R. et al. (2006) Eur. J. Neurosci. 24, 94.
- Vertes, R.P. and Kocsis, B. (1997) Neuroscience 81, 893.
- Wechselberger, M. et al. (2006) Am. J. Physiol. 291, R518.
- Whitaker, G.M. et al. (2007) J. Biol. Chem. 282, 22900.
- Yano, S. et al. (2008) Biomed. Res. 29, 195.
- Ye, B. and Nerbonne, J.M. (2009) J. Biol. Chem. 284, 25553.
- Ying, S.W. et al. (2007) J. Neurosci. 27, 8719.
- Zhang, Q. et al. (2009) Biochim. Biophys. Acta. 1788, 1138.
- Zicha, S. et al. (2005) Cardiovasc. Res. 66, 472.
- Zolles, G. et al. (2009) Neuron 62, 814.