L-type voltage-sensitive Ca2+ channels (CaV) on the plasma membrane of diverse cell types play a fundamental role in Ca2+ signaling and cellular function. In many cases, CaV channel signals are associated with gene transcription. This signal has distinguishing properties that include site of entry subcellular localization, amplitude and duration. This mini review briefly describes how regulation of the Ca2+ signal by CaV1 is used to transform specific firing patterns into qualitatively and quantitatively distinct nuclear functions.
Introduction
Many types of Ca2+ channels have been described in both the nervous system and peripheral tissues such as endocrine, skeletal, cardiac, and smooth muscle (Fig. 5-7). The CaV1 channel couples membrane depolarization to cell surface receptor stimuli in order to regulate a multitude of processes including gene expression, synaptic efficacy, mRNA stability, and cell survival. They are exclusively expressed in excitable cells such as skeletal, cardiac, smooth muscle, neurons and endocrine cells.1-6 Increases in intracellular Ca2+ concentration is induced by electrical or receptor stimuli caused by CaV1 activation often occurs as repetitive Ca2+ spikes or Ca2+ oscillations. The parameters of Ca2+ entry such as amplitude, frequency, intracellular location and speed of Ca2+ waves are regulated by the type and the intensity of the stimulus in the same cell.7,8 These distinct modes of Ca2+ entry into a cell play a key role in exterminating which signaling pathways are activated and thus specify the cellular response and induced selective cellular function.9
Ca2+ Initiates Transcription
Cells mainly employ two strategies to couple Ca2+ stimulus to transcription. First, Ca2+ (at the site of entry), activates cytoplasmic signaling molecules which conveys the signal to the nucleus. These signals can be blocked by the use of CaV1-specific inhibitors (FS-2 (#F-700), Calciseptine (#C-500) and Calcicludine (#C-650, replaced with #SPC-650)). In addition, the CaV1 activator (±)-Bay K8644 (#B-350), has been shown to modulate the extracellular signal regulated by the serine/threonine kinase (ERK)/mitogen-activated protein kinase (P42/44 MAPK) cascade in many cell types (Figs. 1, 2). This signaling pathway plays a fundamental role in cell proliferation, differentiation, motility, survival and apoptosis by transcription dependent and independent mechanisms in a variety of cell types.10-12 For example, in human islets, glucose induced Ca2+ over stimulation is responsible for β cell dysfunction and induction of apoptosis (Fig. 2).13
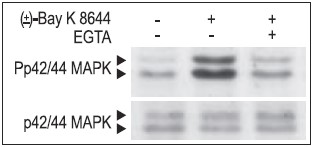
Jurkat T cells were preincubated with or without 2 mM EGTA to chelate Ca2+ for 20 min, then stimulated with 50 μM (±)-Bay K8644 (#B-350) for 10 min. The cell proteins were resolved by SDS PAGE, probed with Anti-Phospho-p42/44 MAPK (upper panel) or with Anti-p42/44 MAPK (lower panel).
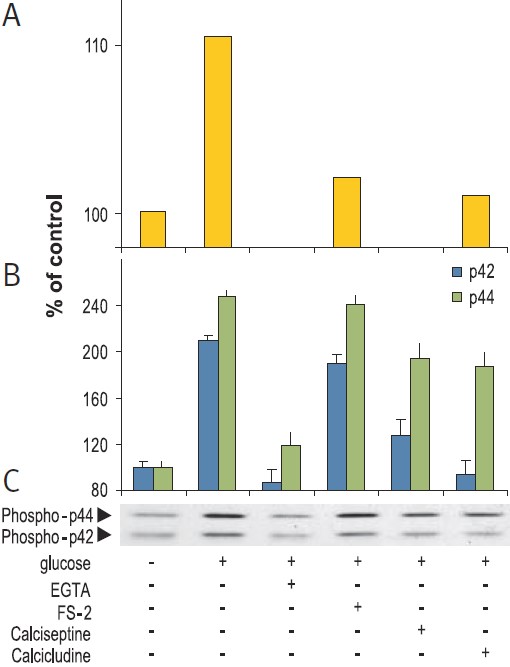
RIN beta cells were stimulation for 15 min with 20 mM glucose in presence or absence of 5 mM EGTA, 2 μM FS-2 (#F-700), 10 μM Calcicludine (#C-650, replaced with #SPC-650) or 10 nM Calciseptine (#C-500). Graph A presents the intracellular levels of Ca2+, 10 sec post stimulation, in Fura-2 AM loaded cells. Picture C shows the western blot analysis of active p42/44 MAP Kinase probed with an anti-phospho-p42/44 MAPK and graph B shows the intensity of p42 and p44 bands.
Secondly, Ca2+ can act directly in the nucleus. For instance, activation of CaV1 and Ca2+ influx has been shown to activate the transcription factor Cyclic Adenosine 3’,5’-Monophosphate Response Element Binding protein (CREB) (Fig 3).14 This transcription factor regulates the expression of immediate early genes15, and is thought to be important in the formation of long-term memory in the brain.16
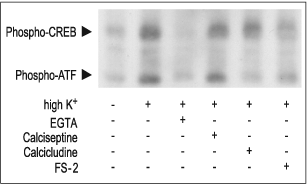
PC12 cells stimulated for 10 min with 35 mM High K+ in presence or absence of 5 mM EGTA, 2 μM FS-2 (#F-700), 10 μM Calcicludine (#C-650, replaced with #SPC-650) or 10 nM Calciseptine (#C-500). The cell extracts were blotted and probed with anti-phospho-CREB which also recognized phospho-ATF.
Calmodulin Mediates the Ca2+ Signal to the Nucleus
The intracellular receptor for Ca2+ is calmodulin. This protein is a very sensitive sensor of Ca2+ concentration, capable of discrimination between signals that differ in spike frequency, amplitude and duration, through its effects on a variety of calmodulin-binding proteins. Changes in intracellular Ca2+ concentration affect calmodulin in three distinct ways: by altering its subcellular distribution; by directing a variety of conformational states of calmodulin that result in target-specific activation and by creating different modes of association with many target proteins.17
Binding of Ca2+ to calmodulin allows the cell to link Ca2+ changes with phosphorylation by activation of the multifunctional Ca2+/calmodulin-dependent protein kinase (CaMK, Fig 4). This kinase has the capability to directly anchor to the pore-forming α1C subunit of CaV1. The frequency response and the state activation of CaMK are tightly modulated by Ca2+ and it reflects the frequency and the amplitude of Ca2+ spikes. CaMK has been shown to play a key role in synaptic plasticity, learning and memory and, in the heart has been implicated in the regulation of gene expression.18
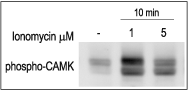
3T3-L1 cells were starved for 2h and then stimulated with 1 or 5 μM Ionomycin (#I-700) for 10 min. The cell extracts were blotted and probed with an antibody for phospho-(Thr268)-CAMKII antibody.
The CaV1 contains two calmodulin binding sites, an “IQ” motif and an upstream calmodulin binding domain – “LA” motif within the carboxy terminus. At resting states, Ca2+-free calmodulin is tethered to the LA motif and lies close to the pore region. During CaV1 activation, when Ca2+ enters through the pore, it is captured by calmodulin. The Ca2+/calmodulin complex switches to the downstream IQ motif, which has a high affinity site for the complex. The Ca2+/calmodulin complex is critical for conveying the signal from the mouth of the CaV1 pore to the nucleus. Binding of Ca2+/calmodulin to the IQ region of CaV1.2 is necessary for its activation and for trafficking of the Ras/P42/44 MAPK pathway, which conveys the signals to the nucleus by activation of transcription factors (Fig. 5).19-21 In the nervous system, this process is essential for neuronal survival, synaptic plasticity and formation of memory.20,22 For instance, in many cell types, voltage activation of CaV1 by K+ depolarization positively stimulates the ERK pathway. In the brain this activation is critical for re-phosphorylation of the synaptic vesicle protein synapsin I and for the activation of several transcription factors.23

Activation of CaV channel and Ca2+ entry through the pore, promotes the binding of calmodulin to Ca2+ and causes calmodulin conformational changes which allow it to migrate downstream from the LA motif to the IQ motif in the COOH terminus. This process in turn regulates the opening of the channel and simultaneously facilitates the activation of signaling pathways.
The transport of Ca2+ to the nucleus is likely to be mediated by its release from internal Ca2+ stores, although it may be a result of the translocation of the Ca2+/calmodulin complex from the cell surface to the nucleus14 (particularly in nerves by retrograde dendritic action potentials) to support activation of CREB, which is strongly associated with dendritic development, neuronal survival and cognition.23,24 The nuclear mobilization of Ca2+/calmodulin is exclusively promoted by CaV1 and glutamate receptor activity,14 but it is modulated by other cellular signals which control calmodulin’s phosphorylation state. The actions of the phosphocalmodulin differ from those of the non-phosphorylated species.25-26
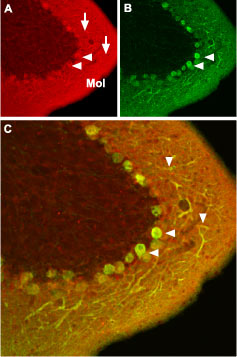
Immunohistochemical staining of CaV1.2 channel with Anti-CaV1.2 (CACNA1C) Antibody (#ACC-003) in mouse cerebellum. (A) CaV1.2 channel (red) appears in Purkinje cells (horizontal arrows) and is distributed diffusely in the molecular layer (Mol) including in Purkinje dendrites (vertical arrows). (B) Staining of Purkinje nerve cells with mouse anti-calcium binding protein antibody (green) in the section demonstrates the location of dendrites in the molecular layer. (C) Confocal merge of CaV1.2 and CBD28K.
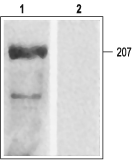
Western blotting of rat ventricular membranes: 1. Anti-CaV1.2a (CACNA1C) Antibody (#ACC-013) (1:200). 2. Anti-CaV1.2a (CACNA1C) Antibody preincubated with the negative control antigen.
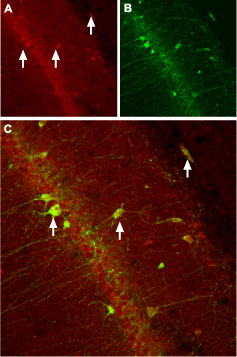
Immunohistochemical staining of CaVβ3 channel with Anti-CACNB3 Antibody (#ACC-008) in rat hippocampus. (A) CaVβ3 channel (red) appears in neurons (arrows). (B) Staining of nerve cells with mouse anti-parvalbumin (a calcium binding protein, green) demonstrates the restriction of CaVβ3 to cell bodies. (C) Confocal merge of CaVβ3 and parvalbumin demonstrates some co-localization of these proteins.
Conclusion
In biological systems the Ca2+ signal serves as an universal intracellular messenger which modulates multiple Ca2+-regulated process. The intracellular mechanisms which decode this Ca2+ signal are complicated and depend upon processes which engage a wide range of enzymatic activity. The CaV1 signal is a unique message which involves intracellular Ca2+ fluctuations together with direct control of Ca2+ binding protein-calmodulin by initiating specific signal transduction pathways and gene transcription.
References
- Ashcroft, F.M. et al. (1994) J. Cell Biochem. 55, 54.
- Bading, H. et al. (1993) Science 260, 181.
- Bean, B.P. (1989) Annu. Rev. Physiol. 51, 367.
- Charles, A.C. et al. (1999) J. Biol. Chem. 274, 7508.
- De Koninck, P. and Cooper E. (1995) J. Neurosci. 15, 7966.
- Christie, B.R. et al. (1997) J. Neurophysiol. 77, 1651.
- Berridge, M.J. et al. (2000) Nat. Rev. Mol. Cell Biol. 1, 11.
- Cruzalegui, F.H. and Bading, H. (2000) Cell. Mol. Life Sci. 57, 402.
- Kiselyov, K. et al. (2003) Cell. Signal. 15, 243.
- Shimamura, A. et al. (2000) Curr. Biol. 10, 127.
- Bonni, A. et al. (1999) Science 286, 1358.
- Ballif, B.A. and Blenis, J. (2001) Cell Growth Differ. 12, 397.
- Maedler, K. et al. (2004) Diabetes 53, 1706.
- Deisseroth, K. et al. (1998) Nature 392, 198.
- Wantanabe, S. et al. (1997) Blood 89, 1197.
- Abel, T. et al. (1998) Science, 279, 338.
- Chin, D. and Means, A.R. (2000) Trends Cell Biol. 10, 322.
- De Koninck, P. and Schulman, H. (1998) Science 279, 227.
- Tebar, F. et al. (2002) Mol. Biol. Cell 13, 2057.
- Dolmetsch, R.E. et al. (2001) Science 294, 333.
- Cullen, P.J. and Lockyer P.J. (2002) Nat. Rev. Mol. Cell Biol. 3, 339.
- Selcher, J.C. et al. (2002) The Neuroscientist 8, 122.
- Cruzalegui, F.H. and Bading, H. (2000) Cell. Mol. Life Sci. 57, 402.
- Gao, L. et al. (2006) J. Neurosci. 26, 6259.
- Nakamura, T. et al. (1999) Neuron 24, 727.
- Benaim, G. and Villalobo, A. (2002) Eur. J. Biochem. 269, 3619.